Custom Split Example#
Contributed by Franz Görlich.
Import core libraries:
from pathlib import Path
import plinder
from plinder.data.splits import (
split,
get_default_config,
)
from plinder.core.scores import query_index
plinder_cfg = plinder.core.get_config()
plinder_local_storage = plinder_cfg.data.plinder_dir
Split Config#
First, let’s modify the split config. We will first get the default config using get_default_config() and then change some of the parameters.
cfg = get_default_config()
print(f'Validation set size: {cfg.split.num_val}')
print(f'Test set size: {cfg.split.num_test}')
print(f'Minimum size of each cluster in the validation set: {cfg.split.min_val_cluster_size}')
Validation set size: 1000
Test set size: 1000
Minimum size of each cluster in the validation set: 30
Since we reduced the total number of samples, let’s also reduce the minimum validation set cluster size, so we avoid removing to many systems.
All configs can be found here
cfg.split.num_test = 500 # Reduce the max size of the test set
cfg.split.num_val = 500 # Reduce the max size of the validation set
cfg.split.min_val_cluster_size = 5 # Reduce the minimum required size of each cluster in the validation set
Custom Dataset#
Let’s generate a custom dataset that we want to resplit. First, let’s load the plindex and then create a custom dataset.
cols_of_interest = [
"system_id",
"entry_pdb_id",
"ligand_ccd_code",
"ligand_binding_affinity",
"ligand_is_proper",
"ligand_molecular_weight",
"system_has_binding_affinity",
]
custom_df = query_index(
columns=cols_of_interest, splits=["train", "val", "test", "removed"]
)
2024-10-25 11:14:50,015 | plinder.core.utils.cpl.download_paths:24 | INFO : runtime succeeded: 0.53s
2024-10-25 11:14:51,601 | plinder.core.utils.cpl.download_paths:24 | INFO : runtime succeeded: 0.55s
custom_df.shape
(567394, 8)
Let’s filter every system out that doesn’t have a binding affinity system_has_binding_affinity, ions and artifacts ligand_is_proper and ligands with a molecular weight of less than 400 g/mol ligand_molecular_weight.
custom_df = custom_df[
(custom_df["ligand_is_proper"] == True) &
(custom_df["system_has_binding_affinity"] == True) &
(custom_df["ligand_molecular_weight"] > 400)
]
custom_df.head(4)
| system_id | entry_pdb_id | ligand_ccd_code | ligand_binding_affinity | ligand_is_proper | ligand_molecular_weight | system_has_binding_affinity | split | |
|---|---|---|---|---|---|---|---|---|
| 20 | 2grt__1__1.A_2.A__1.C | 2grt | GDS | 6.079633 | True | 612.151962 | True | train |
| 22 | 2grt__1__1.A_2.A__2.C | 2grt | GDS | 6.079633 | True | 612.151962 | True | train |
| 74 | 8gr9__1__1.A_1.B__1.C_1.J | 8gr9 | COA | 5.465907 | True | 767.115209 | True | removed |
| 85 | 1grn__1__1.A_1.B__1.C_1.D_1.E | 1grn | GDP | 3.428291 | True | 443.024330 | True | train |
custom_df.shape
(36247, 8)
custom_df['split'].value_counts()
split
train 25610
removed 10483
val 92
test 62
Name: count, dtype: int64
Resplitting the Dataset#
We see that we end up with only 92 systems in our validation set and 62 in our test set. We also have over 10k removed systems. Let’s resplit the dataset and see how the new split looks like.
NOTE: resplitting the dataset requires a lot of memory and might only be feasible on a HPC cluster.
data_dir = Path(plinder_local_storage)
custom_systems = set(custom_df['system_id'].unique())
split_name = 'custom_1'
new_split_df = split(
data_dir=data_dir,
cfg=cfg, # here we use the modified config from earlier
relpath=split_name,
selected_systems=custom_systems
)
new_split_df.shape
(33047, 13)
new_split_df.head(4)
| system_id | uniqueness | split | cluster | cluster_for_val_split | system_pass_validation_criteria | system_pass_statistics_criteria | system_proper_num_ligand_chains | system_proper_num_pocket_residues | system_proper_num_interactions | system_proper_ligand_max_molecular_weight | system_has_binding_affinity | system_has_apo_or_pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10gs__1__1.A_1.B__1.C | 10gs__A_B__C_c101993 | train | c62 | c0 | True | True | 1 | 24 | 15 | 473.162057 | True | False |
| 1 | 10gs__1__1.A_1.B__1.E | 10gs__A_B__E_c101949 | train | c62 | c0 | True | True | 1 | 24 | 13 | 473.162057 | True | False |
| 2 | 19gs__1__1.A_1.B__1.C_1.D | 19gs__A_B__C_D_c147080 | train | c62 | c0 | False | True | 2 | 30 | 12 | 787.630334 | True | False |
| 3 | 19gs__1__1.A_1.B__1.F_1.G | 19gs__A_B__F_G_c101954 | train | c62 | c0 | False | True | 2 | 30 | 14 | 787.630334 | True | False |
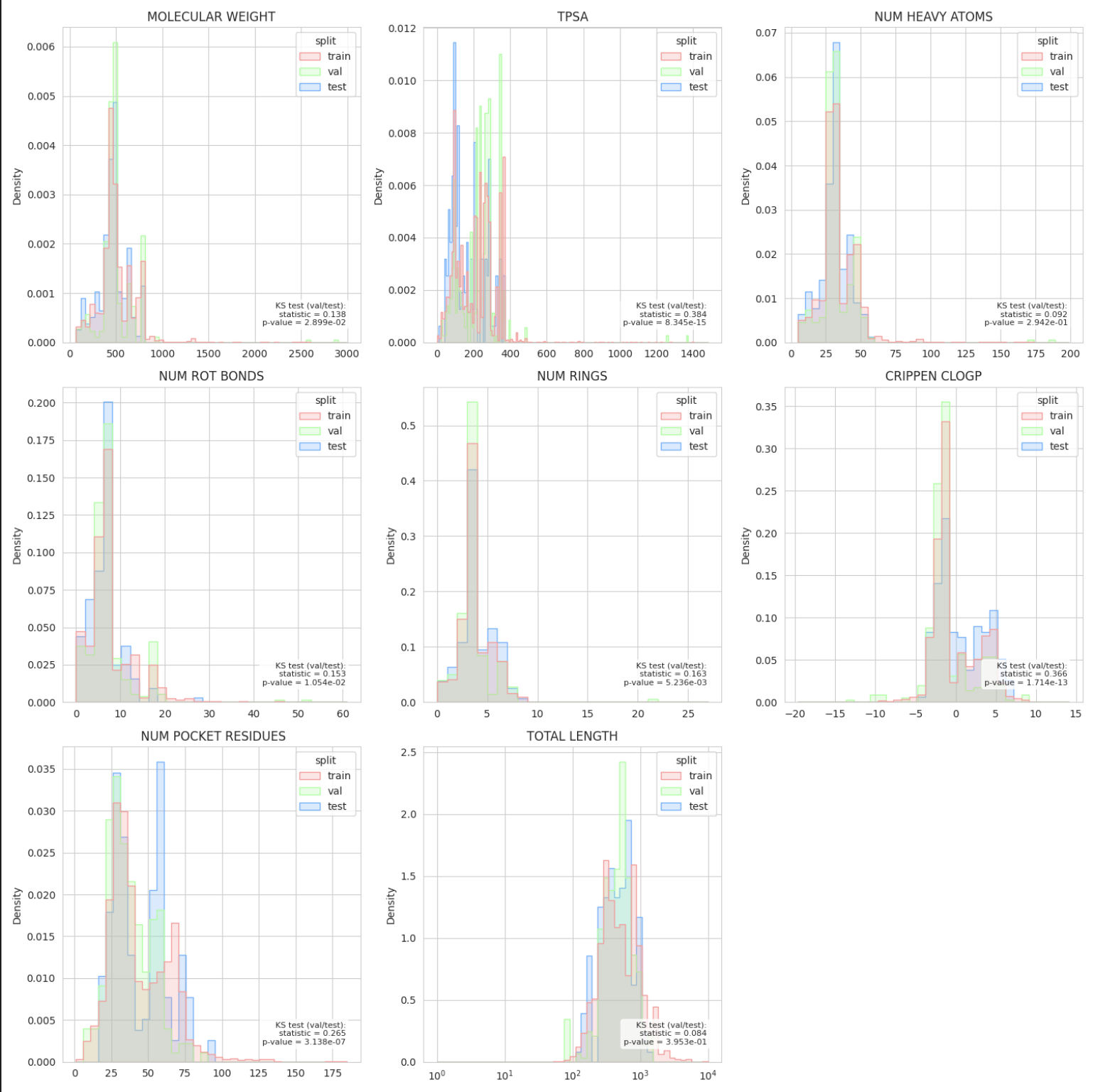
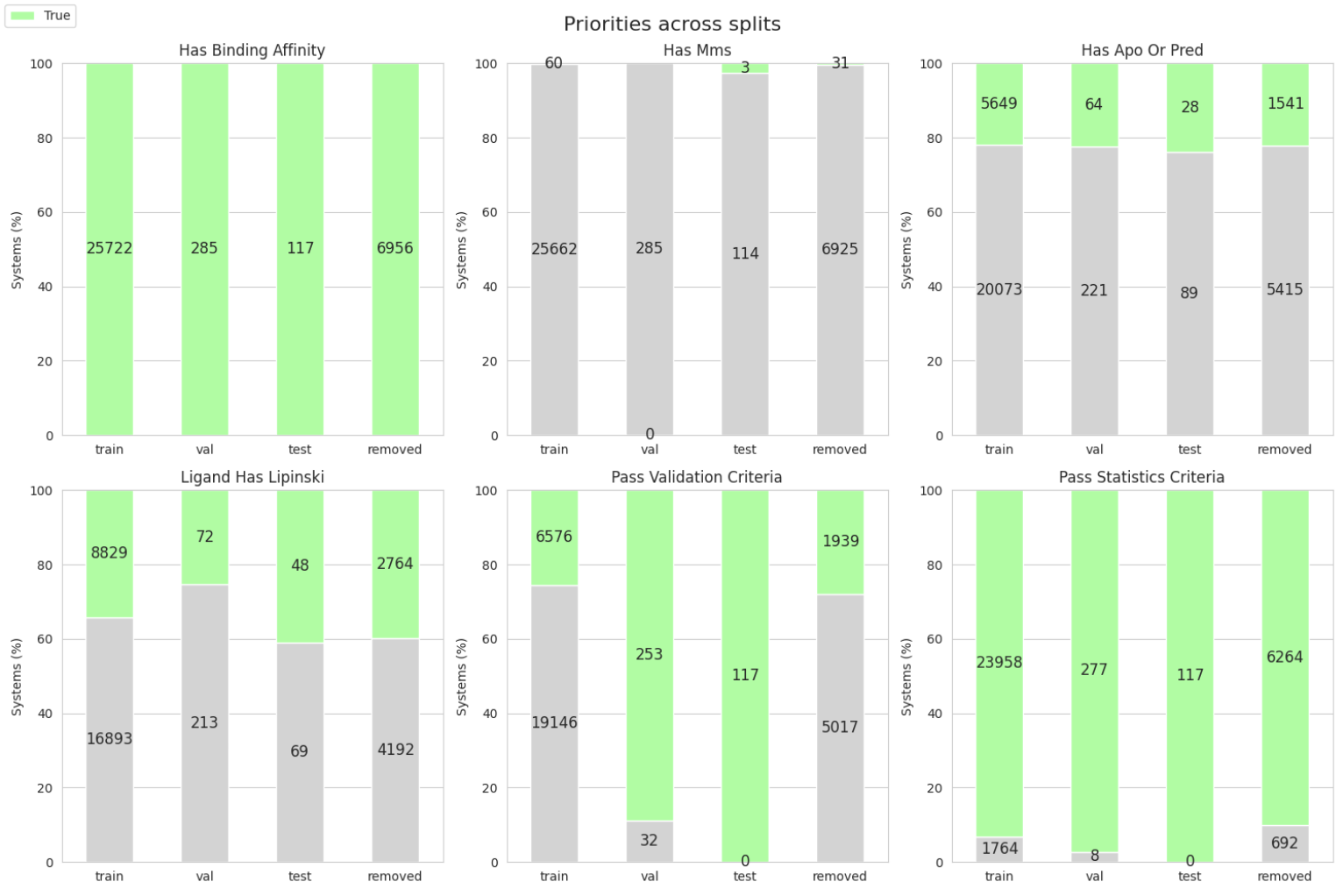
Visualizing the new Split#
Now that we have our first custom split, let’s use the SplitPropertiesPlotter to visualize the new split.
from plinder.core.split.plot import SplitPropertiesPlotter
plotter = SplitPropertiesPlotter.from_files(
data_dir = Path(plinder_local_storage),
split_file = Path(f'{plinder_local_storage}/splits/split_{split_name}.parquet'),
)
This will create a folder split_plots in the current working directory with the following plots:
split_plots/split_proportions.png
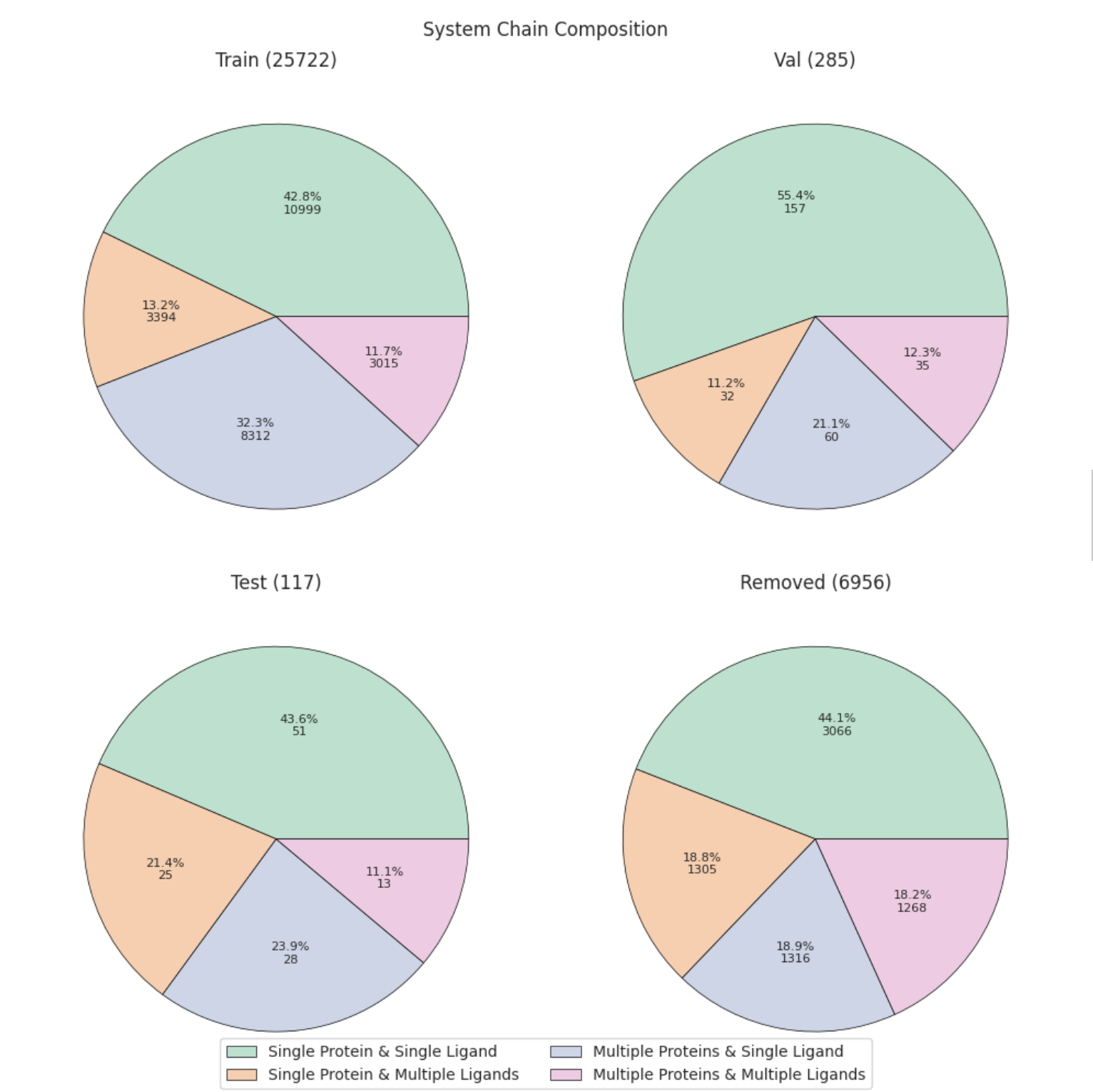
split_plots/chain_composition.png

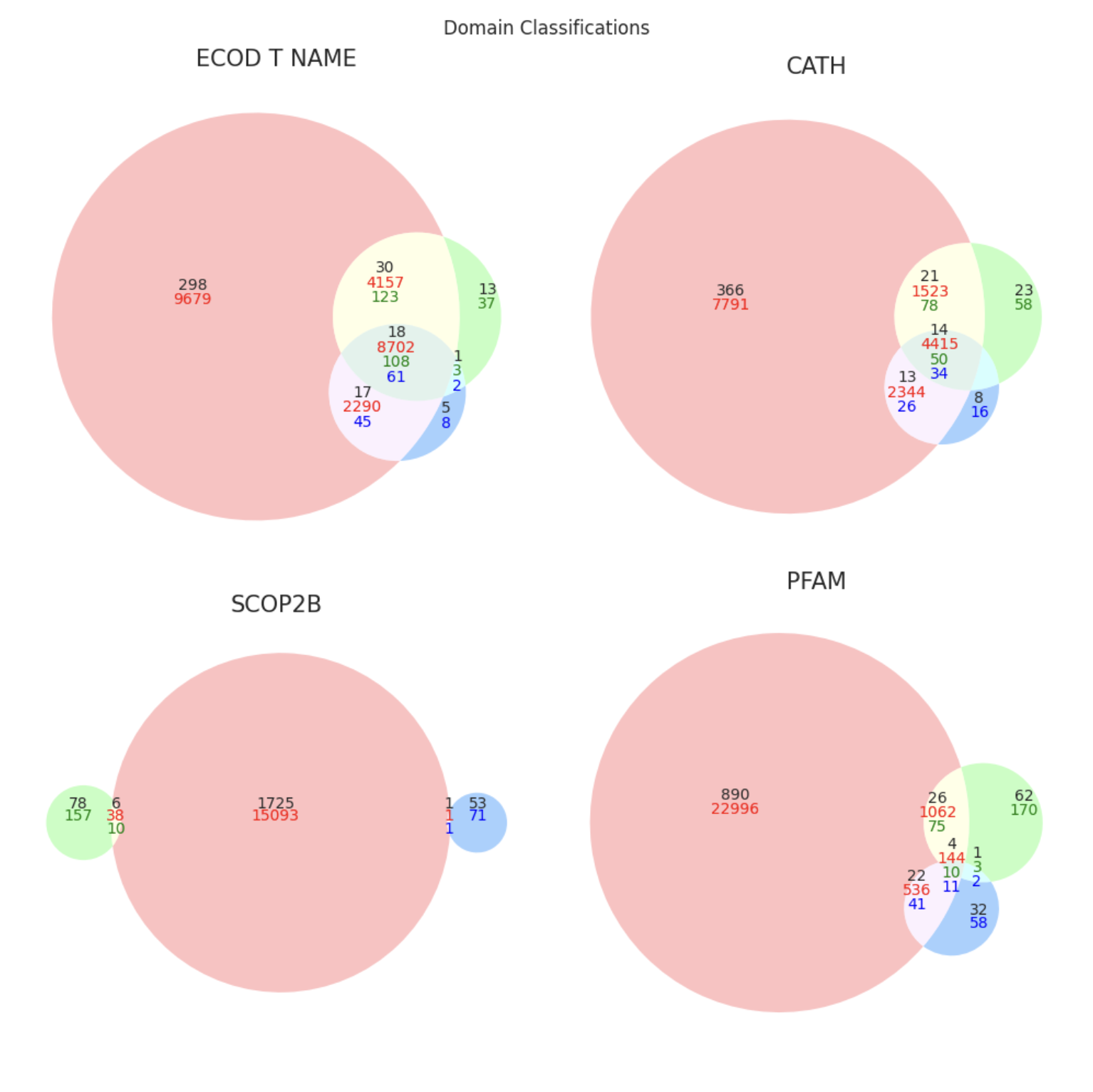
split_plots/domain_classifications.png
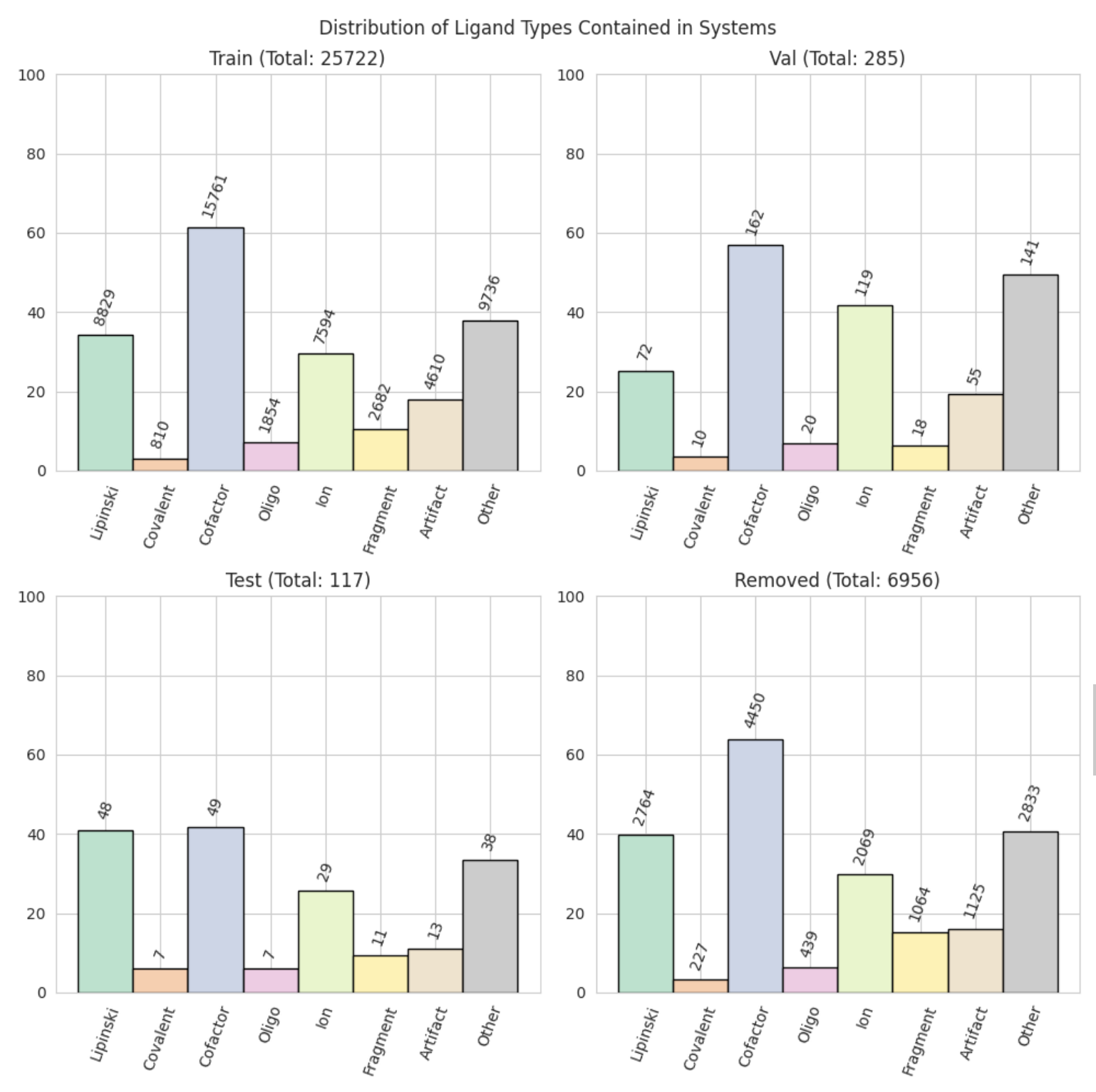
split_plots/ligand_types.png
split_plots/molecular_descriptors.png
split_plots/priorities.png
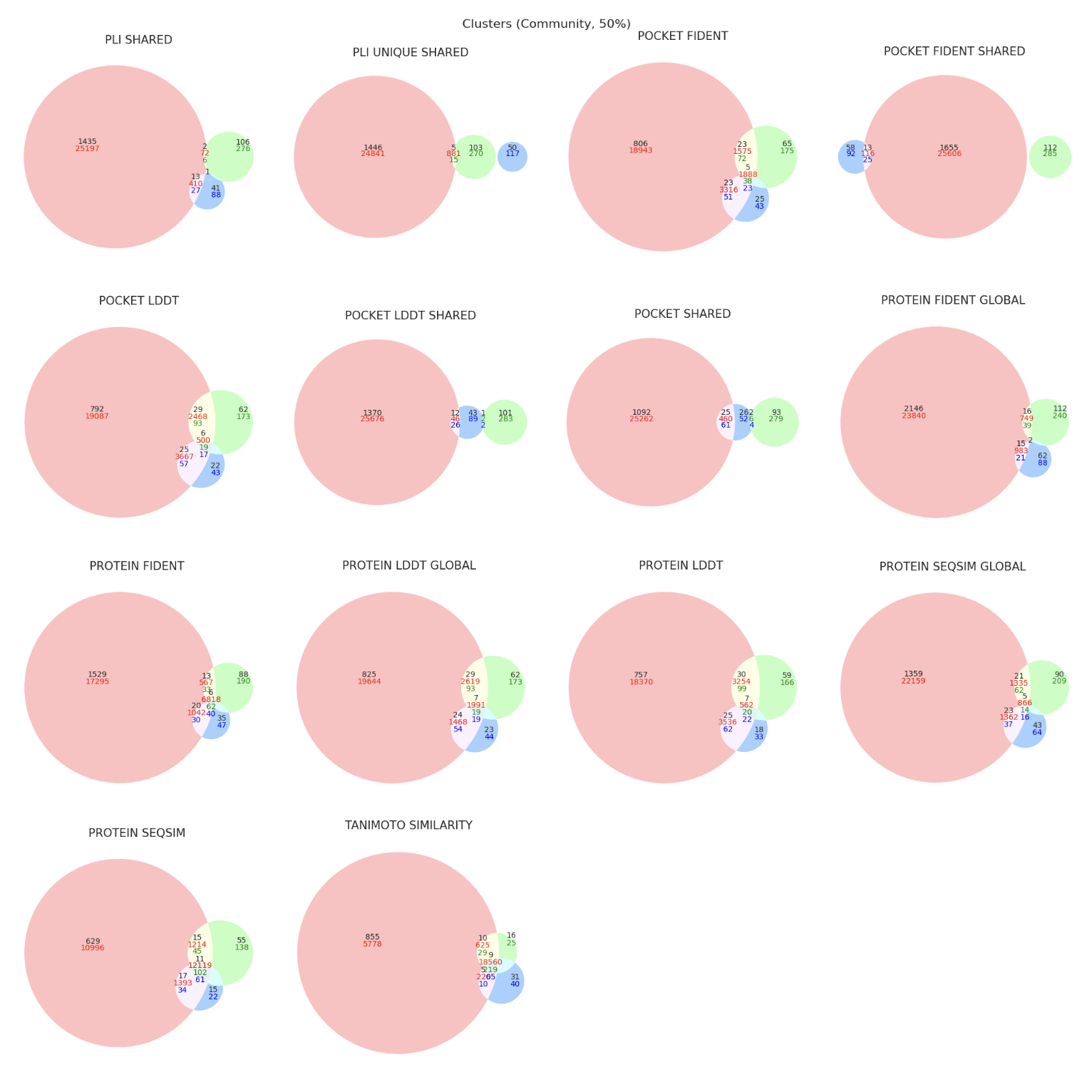
split_plots/plinder_clusters.png